Naef lab – Laboratory of Computational and Systems Biology
Current Research Areas
Our lab is interested in quantitative systems biology and more generally the link between physics and biology. We work on various problems including circadian rhythms, transcriptional bursting, developmental patterning, gene regulation, and single cell imaging. To study these systems we combine theoretical, computational and experimental methods.
• Circadian gene regulatory networks in mammals
• Chronobiology of the liver
• Circadian oscillators in single cells
• Interactions of circadian oscillators and cell cycle
• Transcriptional bursting and noise in mammalian gene transcription
Want to join us for your Student’s project, PhD or Post-doc?
See Openings and please contact [email protected]
Don’t forget to follow us @NaefLab
HIGHLIGHTS
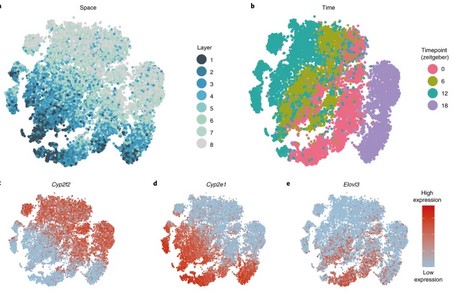
Space-time logic of liver gene expression at sub-lobular scale
The mammalian liver is a central hub for systemic metabolic homeostasis. Liver tissue is spatially structured, with hepatocytes operating in repeating lobules, and sub-lobule zones performing distinct functions. The liver is also subject to extensive temporal regulation, orchestrated by the interplay of the circadian clock, systemic signals and feeding rhythms. However, liver zonation has previously (…)
The circadian oscillator analysed at the single-transcript level
The circadian clock is an endogenous and self-sustained oscillator that anticipates daily environmental cycles. While rhythmic gene expression of circadian genes is well-described in populations of cells, the single-cell mRNA dynamics of multiple core clock genes remain largely unknown. Here we use single-molecule fluorescence in situ hybridisation (smFISH) at multiple time points to measure pairs (…)
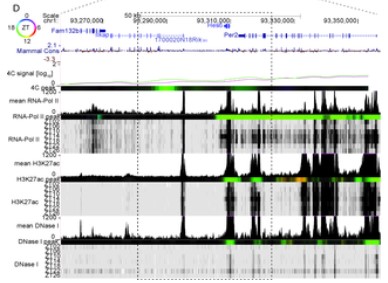
Oscillating and stable genome topologies underlie hepatic physiological rhythms during the circadian cycle
The circadian clock drives extensive temporal gene expression programs controlling daily changes in behavior and physiology. In mouse liver, transcription factors dynamics, chromatin modifications, and RNA Polymerase II (PolII) activity oscillate throughout the 24-hour (24h) day, regulating the rhythmic synthesis of thousands of transcripts. Also, 24h rhythms in gene promoter-enhancer chromatin looping accompany rhythmic mRNA (…)
See all our previous Research Highlights
Contact
Laboratory of Computational and Systems Biology
Monday to Friday: 08.00 – 16.00
Head of laboratory: Prof. Felix Naef
♦ Office AAB 2 48
♦ Phone: +41 21 693 16 21
♦ Email: [email protected]
Administrative Assistant: Sophie Barret
♦ Office AAB 014
♦ Phone: +41 21 693 83 58
♦ Email: [email protected]
Mailing Address:
EPFL SV IBI UPNAE
Prof. Felix Naef
AAB 2 48
Station 15
CH-1015 Lausanne